Print cross-validated eigenvalues
Usage
# S3 method for eigcv
print(x, ...)Arguments
- x
An
eigcvobject created by a call toeigcv().- ...
Ignored.
Examples
library(fastRG)
set.seed(27)
B <- matrix(0.1, 5, 5)
diag(B) <- 0.3
model <- sbm(
n = 1000,
k = 5,
B = B,
expected_degree = 40,
poisson_edges = FALSE,
allow_self_loops = FALSE
)
A <- sample_sparse(model)
eigs<- eigcv(A, k_max = 10)
eigs
#> Estimated graph dimension: 5
#>
#> Number of bootstraps: 10
#> Edge splitting probabaility: 0.1
#> Significance level: 0.05
#>
#> ------------ Summary of Tests ------------
#> k z pvals padj
#> 1 60.0858888 0.000000e+00 0.000000e+00
#> 2 11.7538714 3.372802e-32 3.372802e-32
#> 3 11.1552401 3.375515e-29 3.375515e-29
#> 4 11.3242906 4.974047e-30 4.974047e-30
#> 5 9.5379830 7.281856e-22 7.281856e-22
#> 6 -1.1633387 8.776540e-01 8.776540e-01
#> 7 -1.2996582 9.031409e-01 9.031409e-01
#> 8 -1.1750915 8.800209e-01 8.800209e-01
#> 9 -1.1354378 8.719040e-01 8.719040e-01
#> 10 -0.8694766 8.077067e-01 8.077067e-01
#>
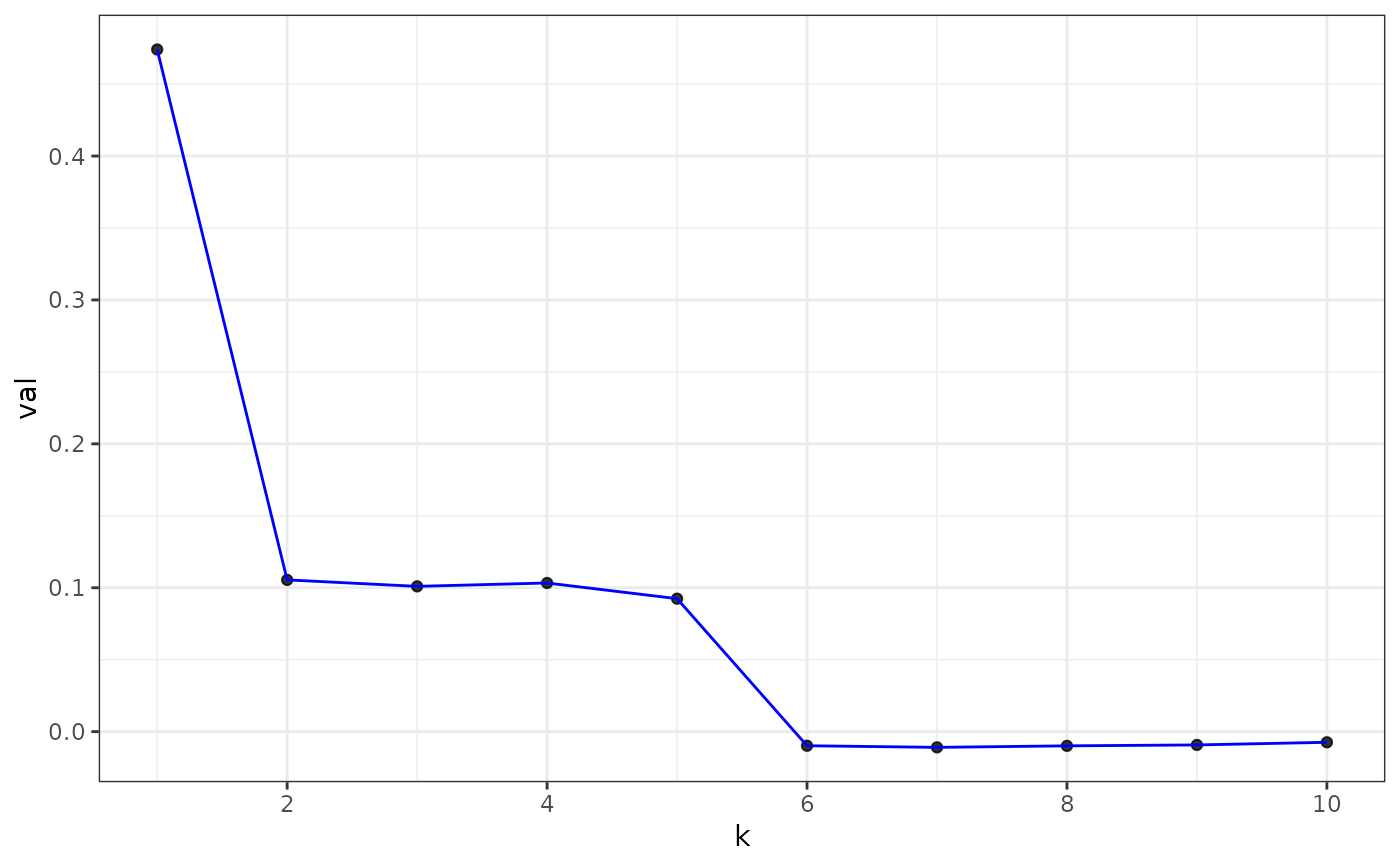
plot(eigs, type = "z-score") # default
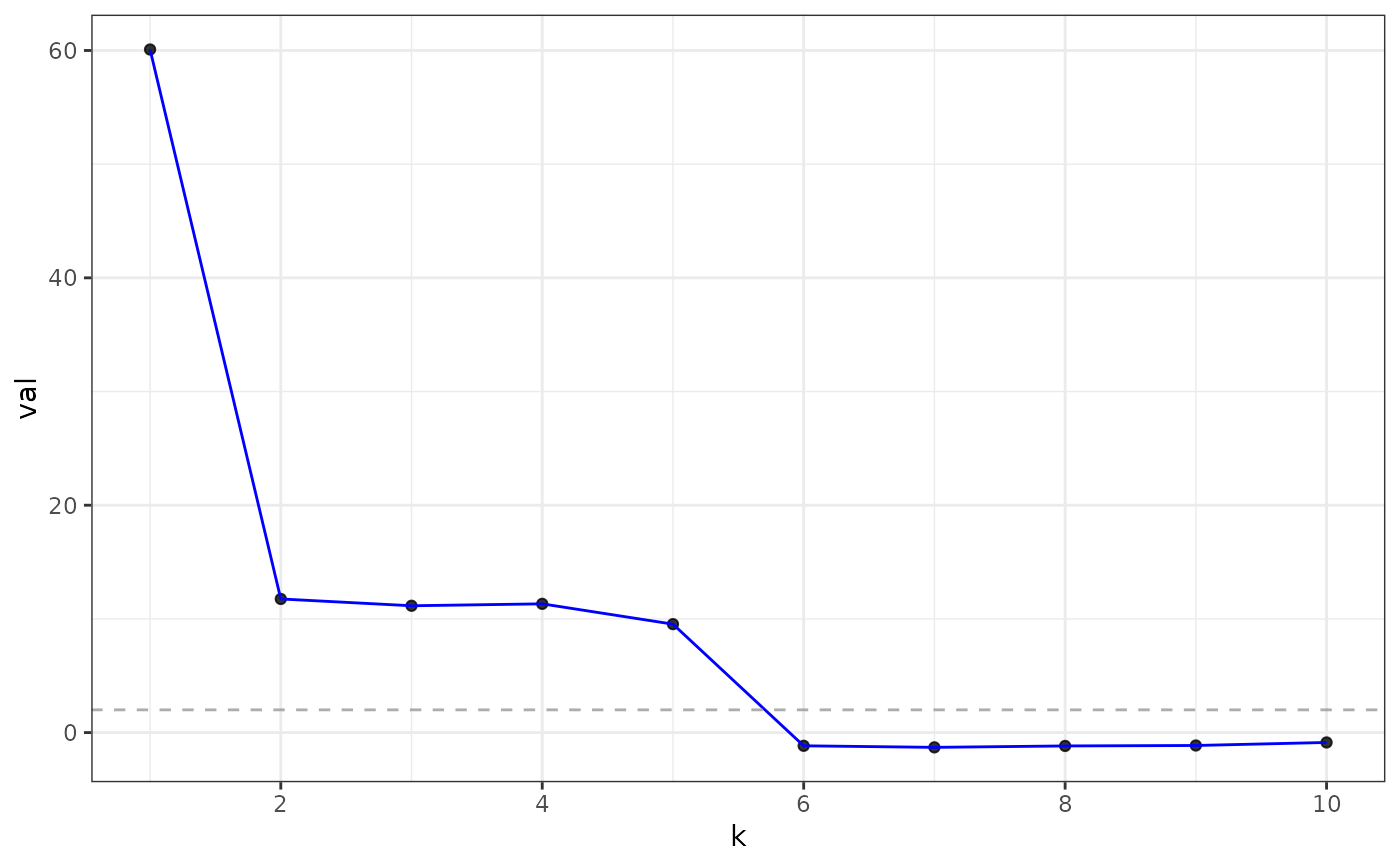 plot(eigs, type = "adjacency")
plot(eigs, type = "adjacency")
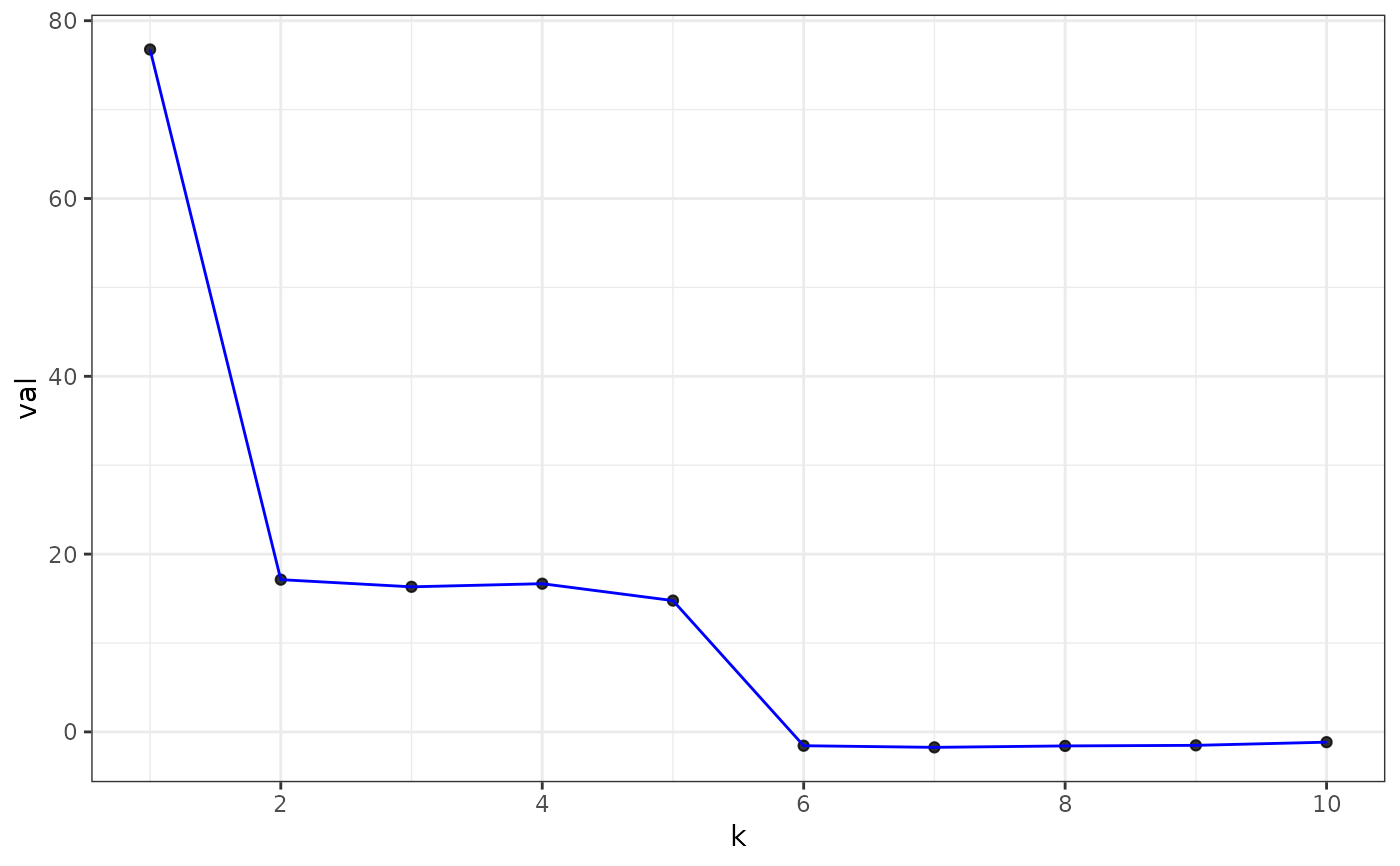 plot(eigs, type = "laplacian")
plot(eigs, type = "laplacian")